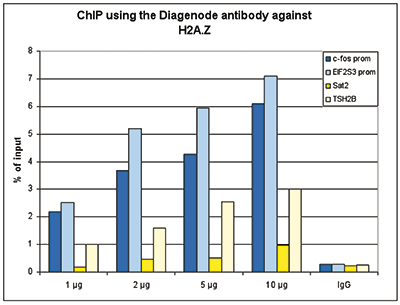
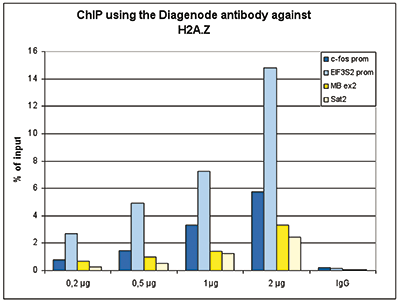
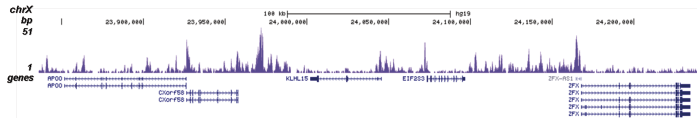
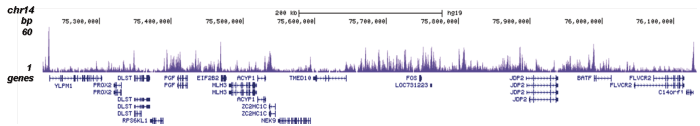
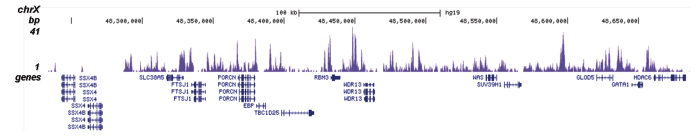
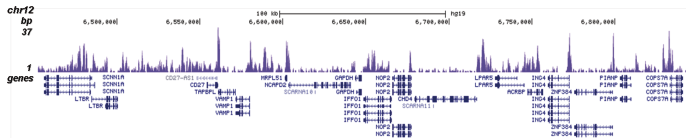
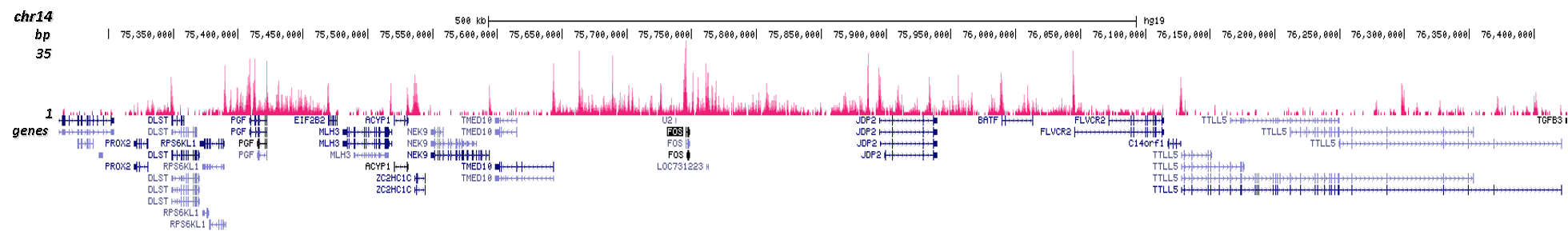
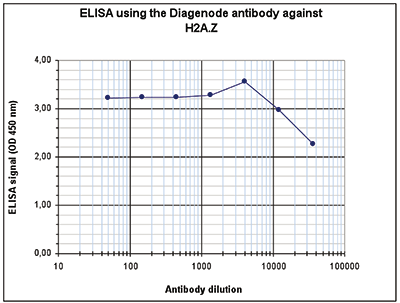
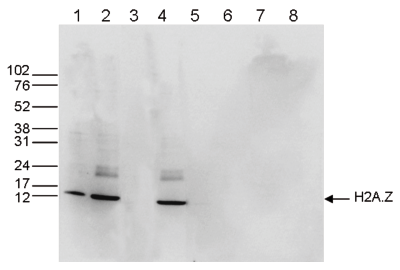
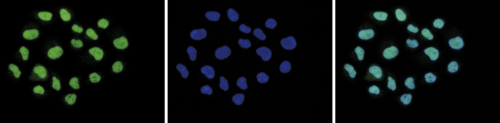
Figure 1. ChIP results obtained with the Diagenode antibody directed against H2A.Z
Figure 1A ChIP assays were performed using human HeLa cells, the Diagenode antibody against H2A.Z (cat. No. C15410201) and optimized PCR primer pairs for qPCR. ChIP was performed with the ““Auto Histone ChIP-seq” kit (cat. No. AB-Auto02-A100) on the IP-Star automated system, using sheared chromatin from 1,000,000 cells. A titration consisting of 1, 2, 5 and 10 μg of antibody per ChIP experiment was analyzed. IgG (2 μg/IP) was used as a negative IP control. Quantitative PCR was performed with primers specific for the promoter of the active genes c-fos and EIF2S3, used as positive controls, and for the inactive TSH2B gene and the Sat2 satellite repeat, used as negative controls. Figure 1B ChIP assays were performed using human K562 cells, the Diagenode antibody against H2A.Z (cat. No. C15410201) and optimized PCR primer sets for qPCR. ChIP was performed with the “iDeal ChIP- seq” kit (cat. No. AB-001-0024) on sheared chromatin from 100,000 cells. A titration of the antibody consisting of 0.2, 0.5, 1 and 2 μg per ChIP experiment was analysed. IgG (1 μg/IP) was used as negative IP control. Quantitative PCR was performed with primers specific for the promoter of the active genes c-fos and EIF2S3, used as positive controls, and for the coding region of the inactive MB gene and the Sat2 satellite repeat, used as negative controls. Figure 1 shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).